What Happens To Mrna After It Completes Transcription
Juapaving
Mar 26, 2025 · 6 min read
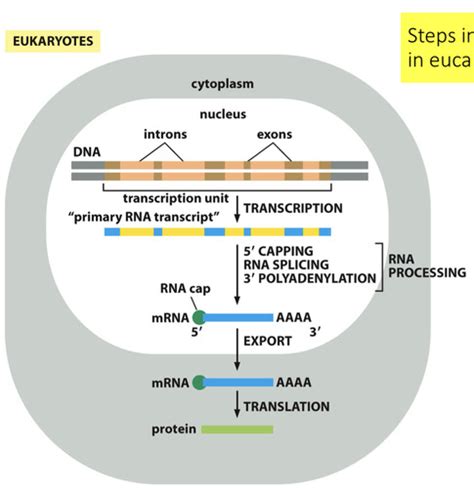
Table of Contents
What Happens to mRNA After It Completes Transcription?
The journey of a messenger RNA (mRNA) molecule is a fascinating odyssey from its creation in the nucleus to its ultimate degradation in the cytoplasm. While transcription marks a crucial step in gene expression, the fate of the newly synthesized mRNA transcript is far from determined. Many intricate processes ensure the accuracy, efficiency, and regulation of protein synthesis, all impacting the lifespan and functionality of the mRNA molecule. This comprehensive guide explores the complex post-transcriptional events that dictate the life and death of mRNA.
From Gene to mRNA: A Recap of Transcription
Before delving into post-transcriptional events, let's briefly revisit the process of transcription. Transcription is the first step in gene expression, where the genetic information encoded in DNA is copied into a complementary mRNA molecule. This process occurs within the nucleus of eukaryotic cells and involves the enzyme RNA polymerase II. RNA polymerase II binds to specific DNA regions called promoters, initiating the unwinding of the DNA double helix and the synthesis of a pre-mRNA molecule.
This pre-mRNA molecule is still an immature form of mRNA. It contains both coding sequences (exons) and non-coding sequences (introns). The next crucial step, therefore, involves processing this pre-mRNA molecule to create a mature, translatable mRNA ready to direct protein synthesis.
Post-Transcriptional Modifications: Maturation of mRNA
The newly synthesized pre-mRNA undergoes several essential modifications before it can be exported from the nucleus and translated into a protein. These modifications are crucial for ensuring the stability, efficiency, and accuracy of protein synthesis. They include:
1. Capping: Protecting the 5' End
The 5' end of the pre-mRNA molecule receives a 5' cap, a 7-methylguanosine (m7G) residue linked to the first nucleotide via an unusual 5'-5' triphosphate linkage. This 5' cap serves several vital functions:
- Protection from Degradation: The cap protects the mRNA from degradation by exonucleases, enzymes that degrade RNA from the ends.
- Initiation of Translation: The cap facilitates the binding of the mRNA to the ribosome, initiating the process of translation.
- Splicing Efficiency: The cap contributes to the efficiency of pre-mRNA splicing.
2. Splicing: Removing Introns
Pre-mRNA molecules contain both exons (coding sequences) and introns (non-coding sequences). Splicing is the process of removing introns and joining exons to create a continuous coding sequence. This process is essential because introns, if left in the mRNA, would lead to the production of non-functional proteins. Splicing is catalyzed by a large ribonucleoprotein complex called the spliceosome, which recognizes specific sequences at the intron-exon boundaries.
Alternative Splicing: A remarkable feature of splicing is its ability to produce multiple mRNA isoforms from a single gene through alternative splicing. This process involves the inclusion or exclusion of different exons during splicing, creating a diversity of protein isoforms with potentially different functions. This greatly expands the proteome, the entire set of proteins expressed by a genome.
3. Polyadenylation: Stabilizing the 3' End
The 3' end of the pre-mRNA undergoes polyadenylation, the addition of a poly(A) tail, a long string of adenine nucleotides (typically 200-250). This poly(A) tail is crucial for:
- mRNA Stability: The poly(A) tail enhances the stability of the mRNA molecule, protecting it from degradation by exonucleases.
- Nuclear Export: The poly(A) tail is recognized by specific proteins that facilitate the export of the mature mRNA from the nucleus to the cytoplasm.
- Translation Initiation: The poly(A) tail contributes to the efficiency of translation initiation.
mRNA Export from the Nucleus
Once the pre-mRNA molecule has undergone capping, splicing, and polyadenylation, it is ready to be exported from the nucleus to the cytoplasm, where protein synthesis takes place. This export process is tightly regulated and involves several nuclear transport factors that bind to the mature mRNA, recognizing its processed 5' cap and poly(A) tail. These factors escort the mRNA through the nuclear pores into the cytoplasm.
mRNA Translation and Protein Synthesis
In the cytoplasm, the mature mRNA molecule encounters ribosomes, the protein synthesis machinery. The ribosome binds to the 5' cap of the mRNA and begins to translate the mRNA sequence into a polypeptide chain. This process involves:
- Initiation: The ribosome binds to the mRNA, recognizing the start codon (AUG).
- Elongation: The ribosome moves along the mRNA, decoding each codon and adding the corresponding amino acid to the growing polypeptide chain.
- Termination: The ribosome encounters a stop codon (UAA, UAG, or UGA), signaling the end of translation. The completed polypeptide chain is released, and the ribosome dissociates from the mRNA.
mRNA Degradation: The End of the Line
mRNA molecules don't exist indefinitely. Their lifespan is regulated and plays a crucial role in controlling gene expression. Several mechanisms contribute to mRNA degradation:
- Deadenylation: The shortening of the poly(A) tail is a crucial initial step in mRNA degradation. Enzymes called deadenylases remove adenine nucleotides from the poly(A) tail, reducing its protective effect and making the mRNA susceptible to degradation.
- Decapping: Once the poly(A) tail is shortened sufficiently, the 5' cap can be removed by decapping enzymes. The removal of the cap exposes the mRNA to degradation from the 5' end.
- Exonucleolytic Degradation: Exonucleases degrade the mRNA from either the 5' or 3' end, progressively shortening the molecule until it's completely degraded.
- Endonucleolytic Cleavage: Some mRNAs undergo endonucleolytic cleavage, where the molecule is cut in the middle by specific enzymes. This can be targeted to specific sequences or triggered by certain cellular conditions.
- Nonsense-Mediated Decay (NMD): NMD is a quality control mechanism that degrades mRNAs containing premature stop codons. These premature stop codons can arise from mutations or errors in splicing, and NMD prevents the production of truncated, potentially harmful proteins.
- RNA Interference (RNAi): RNAi is a powerful mechanism for regulating gene expression through the degradation of specific mRNAs. This process involves small interfering RNAs (siRNAs) or microRNAs (miRNAs) that bind to complementary sequences on the target mRNA, leading to its degradation or translational repression.
Factors Influencing mRNA Lifespan
The lifespan of an mRNA molecule is not uniform; it varies depending on several factors:
- Sequence-Specific Determinants: Specific sequences within the mRNA molecule can influence its stability and susceptibility to degradation.
- RNA-Binding Proteins: Numerous RNA-binding proteins can bind to mRNA molecules, affecting their stability, localization, and translation efficiency. Some proteins stabilize the mRNA, while others promote its degradation.
- Cellular Conditions: Cellular stress, nutrient availability, and other environmental factors can influence mRNA stability and turnover.
Conclusion: A Dynamic Process
The life of an mRNA molecule, from its synthesis in the nucleus to its eventual degradation in the cytoplasm, is a dynamic and tightly regulated process. Post-transcriptional modifications, export, translation, and degradation are all intricately interconnected and contribute to the precise control of gene expression. Understanding these processes is essential for comprehending the complexity of cellular function and for developing therapeutic strategies targeting gene expression. The intricate balance of these mechanisms ensures the accurate, efficient, and timely production of proteins, essential for the life and function of all organisms. Further research continually uncovers new nuances within these pathways, highlighting the ongoing dynamism and importance of mRNA biology.
Latest Posts
Latest Posts
-
What Is A Common Multiple Of 3 4 And 5
Mar 28, 2025
-
31 Inches Is How Many Feet
Mar 28, 2025
-
Is Clockwise Moment Positive Or Negative
Mar 28, 2025
-
44 As A Product Of Prime Factors
Mar 28, 2025
-
A Chord That Passes Through The Center Of The Circle
Mar 28, 2025
Related Post
Thank you for visiting our website which covers about What Happens To Mrna After It Completes Transcription . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
