Forward Primer And Reverse Primer In Pcr
Juapaving
Mar 31, 2025 · 7 min read
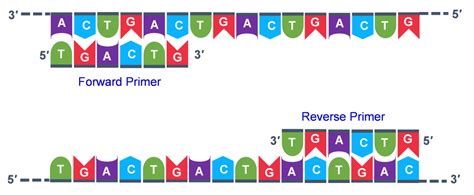
Table of Contents
Forward and Reverse Primers in PCR: A Deep Dive
Polymerase Chain Reaction (PCR) is a cornerstone technique in molecular biology, allowing for the exponential amplification of specific DNA sequences. This process relies heavily on short, single-stranded DNA sequences known as primers. These primers are crucial for initiating DNA synthesis and defining the target region to be amplified. Understanding the role of forward and reverse primers is essential for successful PCR. This article will delve into the intricacies of these primers, exploring their design, functionality, and importance in various PCR applications.
Understanding the Basics: What are Primers?
Primers are short, synthetic oligonucleotides, typically ranging from 18 to 30 base pairs (bp) in length. They act as starting points for DNA polymerase, the enzyme responsible for synthesizing new DNA strands. During PCR, the primers anneal (bind) to complementary sequences on the template DNA, providing a 3'-hydroxyl group (-OH) for the polymerase to extend. Without primers, DNA polymerase cannot initiate DNA synthesis. This is because it requires a pre-existing 3'-OH group to add nucleotides to.
The specificity of the primers is paramount. They must bind only to the target DNA sequence and not to other regions of the genome, ensuring that only the desired DNA fragment is amplified. This specificity is achieved through careful primer design, which considers factors like primer length, melting temperature (Tm), GC content, and potential secondary structures.
The Dynamic Duo: Forward and Reverse Primers
In PCR, two primers are always employed: a forward primer and a reverse primer. They work in concert to amplify the target DNA region.
The Forward Primer: Leading the Charge
The forward primer is designed to be complementary to the template DNA strand at the beginning of the target sequence. Its sequence is identical to the 5' to 3' direction of the sense (coding) strand of the target DNA. During PCR, the forward primer binds to the 3' end of the antisense strand, providing the starting point for DNA synthesis in the 5' to 3' direction. This creates a new strand that is complementary to the antisense strand.
Key characteristics of a good forward primer:
- Specificity: It must bind only to the target sequence.
- Appropriate length: Typically 18-30 bp.
- Optimal melting temperature (Tm): Around 55-65°C.
- Balanced GC content: 40-60%.
- Absence of self-complementarity and hairpin structures: To prevent primer dimers and other undesirable secondary structures.
- Absence of runs of the same base: For example, more than three consecutive Gs or Cs.
- Unique sequence: It should be specific to the target region.
The Reverse Primer: Completing the Cycle
The reverse primer is complementary to the opposite strand of the target sequence at its end. Its sequence is the reverse complement of the 3' to 5' direction of the sense strand of the target DNA. During PCR, the reverse primer binds to the 3' end of the sense strand, providing the starting point for DNA synthesis in the 5' to 3' direction. This creates a new strand that is complementary to the sense strand.
Key characteristics of a good reverse primer:
- Specificity: It must bind only to the target sequence.
- Appropriate length: Typically 18-30 bp.
- Optimal melting temperature (Tm): Similar to the forward primer.
- Balanced GC content: 40-60%.
- Absence of self-complementarity and hairpin structures: To prevent primer dimers and other undesirable secondary structures.
- Absence of runs of the same base: For example, more than three consecutive Gs or Cs.
- Unique sequence: It should be specific to the target region.
Primer Design: A Critical Step
Effective PCR hinges on meticulous primer design. Several factors must be carefully considered:
1. Primer Length:
Optimal primer length typically ranges from 18 to 30 base pairs. Shorter primers may lack specificity, while longer primers can lead to reduced annealing efficiency.
2. Melting Temperature (Tm):
The melting temperature (Tm) is the temperature at which 50% of the primers are bound to the template DNA. A well-designed primer typically has a Tm in the range of 55-65°C. The Tm should be similar for both forward and reverse primers to ensure efficient annealing.
3. GC Content:
The GC content (the percentage of guanine and cytosine bases) should be balanced, typically between 40% and 60%. A high GC content can lead to strong binding, while a low GC content can result in weak binding.
4. Avoiding Secondary Structures:
Primers should be designed to avoid self-complementarity and hairpin structures. These secondary structures can hinder primer annealing and reduce PCR efficiency.
5. Primer Dimers:
Primer dimers are formed when two primers anneal to each other rather than to the template DNA. This can significantly reduce PCR efficiency. It's essential to check for potential primer-dimer formation using online tools.
6. Specificity:
The primers must be specific to the target sequence and not bind to other regions of the genome. BLAST searches can be used to verify primer specificity.
The Role of Forward and Reverse Primers in PCR Amplification
The entire PCR process unfolds through repeated cycles, each comprising three major steps:
- Denaturation: The double-stranded DNA template is heated to separate the strands.
- Annealing: The primers bind (anneal) to their complementary sequences on the single-stranded DNA templates. This step relies on the carefully chosen melting temperatures of the primers; the temperature must be low enough for annealing but high enough to prevent non-specific binding. Both the forward and reverse primers bind simultaneously.
- Extension: DNA polymerase extends the primers by adding nucleotides to the 3' end, synthesizing new DNA strands complementary to the template strands. This step uses the annealed primers as starting points.
Each cycle doubles the amount of target DNA. The forward and reverse primers work in tandem; the forward primer initiating synthesis on one strand while the reverse primer initiates synthesis on the complementary strand. As the cycles progress, the target sequence is amplified exponentially. The final PCR product is a double-stranded DNA molecule with the desired length, bounded by the sequences defined by the forward and reverse primers.
Applications of Forward and Reverse Primers Beyond Basic PCR
The principles of forward and reverse primers extend far beyond basic PCR. Their applications encompass a wide range of molecular biology techniques, including:
- Quantitative PCR (qPCR): Used to measure the amount of starting DNA or RNA. Here, the primers are designed for high specificity and efficiency.
- Reverse Transcription PCR (RT-PCR): Used to amplify RNA molecules by first converting them to cDNA using reverse transcriptase. Specific primers target the cDNA.
- Site-directed mutagenesis: Employed to introduce specific mutations into a DNA sequence. The primers contain the desired mutation.
- Genotyping: Used to identify specific alleles or genetic variations. The primers are designed to amplify specific variants.
- Cloning: Used to insert DNA fragments into vectors. Primers are designed to introduce restriction sites or other modification at the target DNA ends.
Troubleshooting PCR: Primer-Related Issues
Several issues can arise during PCR, often related to primer design or quality. These include:
- No amplification: This might indicate primer failure, incorrect annealing temperature, or insufficient DNA template.
- Non-specific amplification: This suggests primers lacking specificity and binding to unintended regions.
- Weak amplification: This could be due to low primer concentration, inefficient primers, or degraded DNA template.
- Primer dimers: These compete with primer binding to the template, reducing the amplification of the target.
Conclusion: The Unsung Heroes of Molecular Biology
Forward and reverse primers are fundamental components of PCR, providing the essential framework for DNA amplification. Their design and functionality significantly impact the success of the PCR reaction and related techniques. A thorough understanding of primer design principles and their role in amplification is crucial for researchers working in molecular biology, ensuring accurate, reliable results across diverse applications. The precise design of these primers is essential not only for successful amplification but also for the broader field of molecular diagnostics, genetic engineering, and biomedical research. Mastering primer design is a key skill for anyone working in this field, directly impacting the reliability and interpretation of experimental outcomes.
Latest Posts
Latest Posts
-
How Many Neutrons Are In Potassium
Apr 02, 2025
-
Relation Between Linear And Angular Acceleration
Apr 02, 2025
-
What Is The Difference Between Lonely And Alone
Apr 02, 2025
-
What Is The Prime Factors Of 150
Apr 02, 2025
-
Hydrolysis Of Adp Produces Which Of The Following Products
Apr 02, 2025
Related Post
Thank you for visiting our website which covers about Forward Primer And Reverse Primer In Pcr . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
