Difference Between Prokaryotic And Eukaryotic Transcription
Juapaving
Mar 26, 2025 · 6 min read
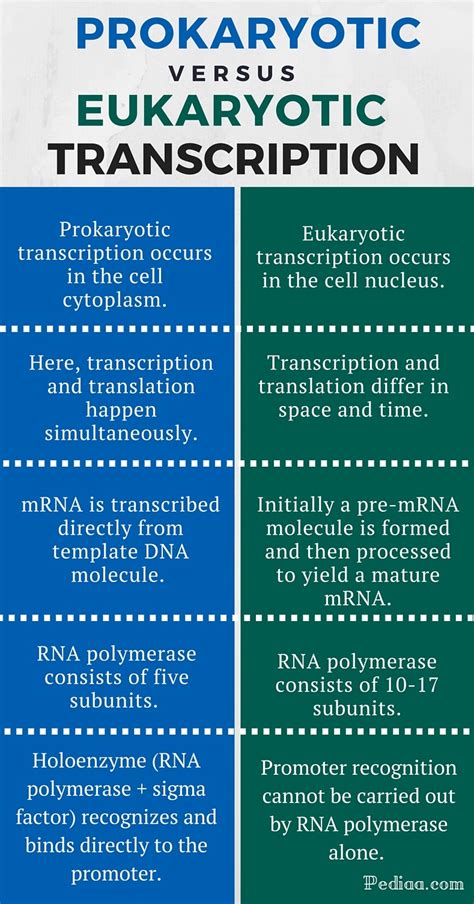
Table of Contents
Delving Deep into the Differences: Prokaryotic vs. Eukaryotic Transcription
Transcription, the fundamental process of converting DNA's genetic code into RNA, is a cornerstone of molecular biology. While the overarching goal remains the same – to synthesize RNA from a DNA template – the mechanisms and intricacies of this process differ significantly between prokaryotes (bacteria and archaea) and eukaryotes (plants, animals, fungi, and protists). Understanding these differences is crucial for comprehending the complexities of gene expression and regulation. This comprehensive article will dissect the key distinctions between prokaryotic and eukaryotic transcription, exploring the players involved, the regulatory nuances, and the ultimate outcomes.
The Transcription Machinery: A Tale of Two Worlds
Both prokaryotic and eukaryotic transcription rely on RNA polymerase, the enzyme responsible for synthesizing RNA molecules. However, the complexity and composition of this enzyme, along with the associated factors, differ considerably.
Prokaryotic Transcription: A Simpler Affair
Prokaryotes, with their simpler cellular structures, possess a relatively straightforward transcription machinery. A single RNA polymerase holoenzyme handles the transcription of all RNA types – mRNA, tRNA, and rRNA. This holoenzyme consists of a core enzyme, composed of five subunits (α2ββ'ω), and a sigma (σ) factor. The sigma factor plays a critical role in promoter recognition, the initial step in transcription. Different sigma factors can recognize different promoter sequences, allowing for the regulation of gene expression under various conditions.
Eukaryotic Transcription: Complexity Takes Center Stage
Eukaryotic transcription presents a far more complex picture. Three distinct RNA polymerases – RNA polymerase I, II, and III – each transcribe different types of RNA:
- RNA polymerase I: Synthesizes ribosomal RNA (rRNA) precursors.
- RNA polymerase II: Transcribes messenger RNA (mRNA) and some small nuclear RNAs (snRNAs).
- RNA polymerase III: Transcribes transfer RNA (tRNA), 5S rRNA, and other small RNAs.
Each polymerase requires a suite of general transcription factors (GTFs) to initiate transcription. These factors bind to the promoter region and recruit the RNA polymerase to the transcription start site. The complexity arises from the diversity of GTFs and their intricate interactions with the promoter and chromatin structure.
Promoter Regions: Setting the Stage for Transcription
The promoter, a DNA sequence upstream of the gene, serves as the binding site for RNA polymerase and other transcription factors. While both prokaryotes and eukaryotes utilize promoters, their structure and the factors that recognize them differ.
Prokaryotic Promoters: Simple yet Effective
Prokaryotic promoters are typically short sequences containing two conserved regions: the -10 region (Pribnow box) and the -35 region. These sequences are located 10 and 35 base pairs upstream of the transcription start site, respectively. The sigma factor recognizes and binds to these regions, facilitating the binding of the core RNA polymerase and initiating transcription. The strength of a promoter, which determines the frequency of transcription initiation, is influenced by the precise sequence of the -10 and -35 regions.
Eukaryotic Promoters: A More Elaborate Affair
Eukaryotic promoters are more diverse and complex than their prokaryotic counterparts. While a core promoter, containing a TATA box (similar in function to the -10 region) and other elements, is essential for transcription initiation, regulatory elements located further upstream or downstream can significantly influence the rate of transcription. These regulatory elements, often referred to as enhancers and silencers, bind transcriptional activators and repressors, respectively, modulating the assembly of the pre-initiation complex and ultimately the transcription rate. Furthermore, the chromatin structure – the packaging of DNA around histone proteins – plays a crucial role in regulating accessibility to the promoter and thus influencing transcription.
Transcription Initiation: The Point of No Return
The initiation of transcription is a tightly regulated process involving multiple steps. The mechanisms and factors involved differ significantly between prokaryotes and eukaryotes.
Prokaryotic Initiation: A Relatively Simple Process
In prokaryotes, the sigma factor within the RNA polymerase holoenzyme plays a central role in initiating transcription. After the sigma factor binds to the promoter, the RNA polymerase unwinds the DNA helix, creating a transcription bubble. The RNA polymerase then synthesizes a short RNA molecule, and the sigma factor is released, allowing the core enzyme to proceed with elongation.
Eukaryotic Initiation: A Multi-Step Orchestration
Eukaryotic transcription initiation is a more elaborate process involving numerous general transcription factors (GTFs). The TATA-binding protein (TBP) within the TFIID complex binds to the TATA box in the core promoter. Other GTFs (TFIIA, TFIIB, TFIIF, TFIIE, TFIIH) then assemble, forming the pre-initiation complex (PIC). TFIIH possesses helicase activity, unwinding the DNA and phosphorylating the C-terminal domain (CTD) of RNA polymerase II, triggering the transition from initiation to elongation. This phosphorylation is crucial for the release of RNA polymerase II from the PIC and its progression along the DNA template.
Transcription Elongation: Building the RNA Molecule
Once transcription initiation is complete, the RNA polymerase proceeds along the DNA template, synthesizing the RNA molecule.
Prokaryotic Elongation: Coupled Transcription and Translation
In prokaryotes, transcription and translation are coupled processes. As the mRNA molecule is synthesized, ribosomes bind to it and begin translating the mRNA into protein. This coupling allows for rapid and efficient gene expression.
Eukaryotic Elongation: Spatial and Temporal Separation
In eukaryotes, transcription and translation are spatially and temporally separated. Transcription occurs in the nucleus, while translation takes place in the cytoplasm. The nascent mRNA molecule undergoes extensive processing before it is exported from the nucleus for translation. This processing includes capping, splicing, and polyadenylation, each essential for mRNA stability and translation efficiency.
Transcription Termination: Bringing the Process to a Halt
The termination of transcription signals the end of RNA synthesis.
Prokaryotic Termination: Rho-Dependent and Rho-Independent Mechanisms
Prokaryotic transcription termination can occur through two main mechanisms: rho-dependent and rho-independent. Rho-dependent termination involves a protein called rho factor, which binds to the RNA transcript and causes the RNA polymerase to dissociate from the DNA template. Rho-independent termination relies on specific DNA sequences within the transcribed region that form hairpin structures in the RNA, causing the RNA polymerase to pause and dissociate.
Eukaryotic Termination: Cleavage and Polyadenylation
Eukaryotic transcription termination is a more complex process, primarily involving cleavage of the RNA transcript at a specific site downstream of the polyadenylation signal (AAUAAA). This cleavage triggers the subsequent polyadenylation of the 3' end of the RNA molecule. The remaining RNA molecule is then degraded, marking the termination of transcription.
Post-Transcriptional Modifications: A Eukaryotic Specialty
Eukaryotic mRNA undergoes extensive post-transcriptional modifications before it can be translated. These modifications are crucial for mRNA stability, export from the nucleus, and translational efficiency. These modifications are largely absent in prokaryotes due to the coupled nature of transcription and translation.
- Capping: Addition of a 7-methylguanosine cap to the 5' end of the mRNA, protecting it from degradation and facilitating ribosome binding.
- Splicing: Removal of introns (non-coding sequences) and joining of exons (coding sequences) to produce a mature mRNA molecule. This process, absent in prokaryotes, significantly increases protein diversity through alternative splicing.
- Polyadenylation: Addition of a poly(A) tail to the 3' end of the mRNA, increasing its stability and influencing translation efficiency.
Conclusion: A Summary of the Key Differences
The differences between prokaryotic and eukaryotic transcription are substantial and reflect the increased complexity of eukaryotic cells. Prokaryotic transcription is a simpler, more streamlined process characterized by a single RNA polymerase, coupled transcription and translation, and relatively simple promoter structures. Eukaryotic transcription, on the other hand, is highly complex, involving multiple RNA polymerases, a multitude of transcription factors, extensive post-transcriptional modifications, and intricate regulation mediated by chromatin structure and regulatory elements. Understanding these differences is crucial for comprehending the diversity and regulation of gene expression in the biological world. Further research into these fascinating processes continues to reveal new insights into the intricacies of life itself.
Latest Posts
Latest Posts
-
Is A Amino Acid A Carbohydrate
Mar 29, 2025
-
The Basic Unit Of Life Is
Mar 29, 2025
-
The Product Of An Objects Mass And Velocity Is Its
Mar 29, 2025
-
The Law Of Segregation Explains That
Mar 29, 2025
-
Diagram Of Animal And Plant Cell
Mar 29, 2025
Related Post
Thank you for visiting our website which covers about Difference Between Prokaryotic And Eukaryotic Transcription . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
